24.02.2014 BiolDiv
From D4Science Wiki
Meeting 24 February 2014, 11:00 am
Skype
Present: Nicolas, Edward
Notes
BiOnym user interfaces
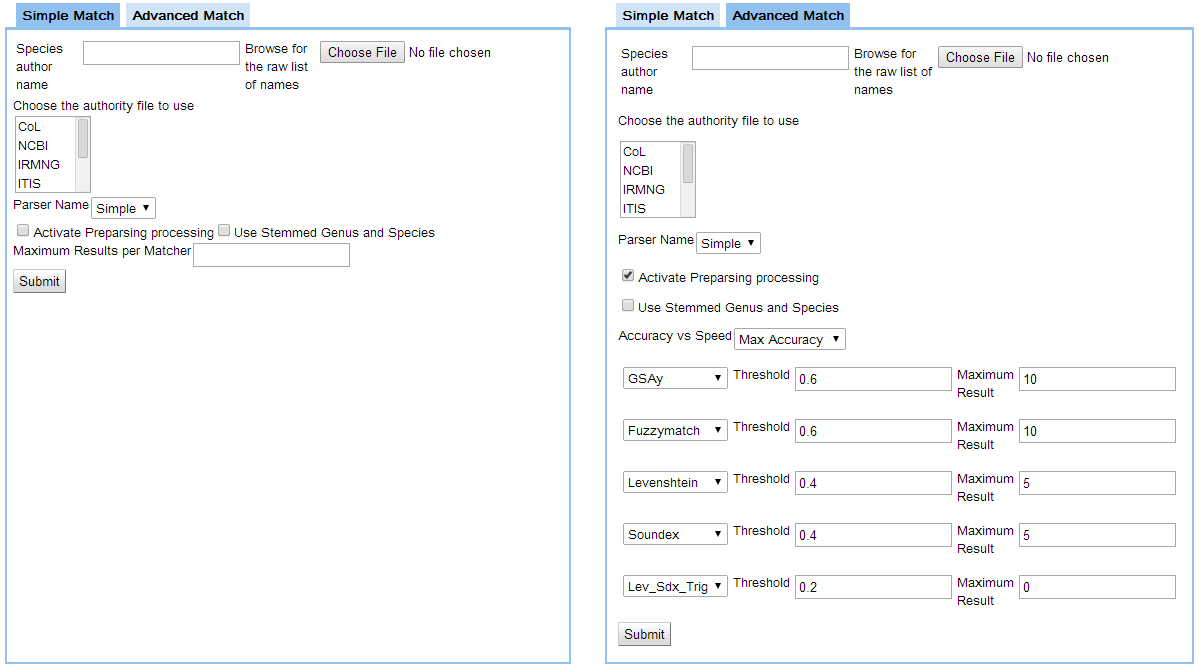
Discussion on the advanced interface sent by Casey just before the discussion - see https://www.dropbox.com/s/7q619udprptxepq/Screenshot%202014-02-24%2018.02.36.png Relevant option will be implemented in the simple one too.
- Replace Labels
- "Species author name" by "Enter one name per line"
- "Browse for the raw list of names" by "Or upload a file"
- "Choose file" by "Browse"
- Suggestion that the upload by default will not ask the user to put the file in the Data Space. If necessary it will be saved there for the analysis purpose, and then removed at the end of the analysis (or the user can be asked at the end of the process when he gets the results). An option as a tick box will allow the user to go to the current upload process, and should be labelled "Check you file consistency" or equivalent. TO DISCUSS next Thursday.
- ASFIS
- It is a TAF for the FAO statistics with a limited number of valid names only. As FishBase/SeaLifeBase are always updated with ASFIS names, it could be more efficient to check against FishBase and to list the ASFIS names in the outputs. TO DISCUSS next Thursday.
- An explanation of what is ASFIS and how to use it should be given on the right of the list box.
- Suggestion to offer an option for a two-steps approach, separating the result of the parser so users can visually inspect the result of the parser. It woul mean to call the workflow without any matcher selection in a first step, the second step being done according to the matcher selection. The default would be one-step workflow. TO DISCUSS next Thursday.
- We need a box to limit the number of results from the matchers altogether. To put on th eright of "accuracy ...".
- Over the five available matcher boxes, fill up by default the first 3 (GSAy, Fuzzy, a third to select from the paper in function of the results of the experimentation)
- We need a tick box to indicate if the authority must be matched or not. Only one for applicable to all selected matchers (if one per matcher, thresholds are not comparable anymore).
- Output selections (as tick boxes):
- TAF IDs: depending on the TAF selection.
- Scientific name
- Authority
- Year: separated from authority (not discussed).
- Classification: if several TAFs, several classification. Limit to Kindgom/Phylum/Class/Order/Family.
- Taxon status: Accepted or Not accepted.
- From WoRMS interface: QualityStatus, Environment and Citation are too variable in the TAF to be taken into account here.
- Should we have a box for the result file name? TO DISCUSS next Thursday.
- Help and User's Guide
- Write a complete document that will be linked from the top
- Extract the mouseover help
- Link to the paper
- Write an explanation show like for AquaMaps (not discussed)
- Other suggestion not discussed TO DISCUSS next Thursday.
- Move Parser name ... on the left of Activate preprocessing ...
- Add the Match authority on the left of Use stemmed ...
Article
- The introduction of Nicolas is more about the need of matching names tools because names are the access key to other species information. Use and cite the recent Patterson's paper in Acta Parasitologica.
- Edward to review the current version first.
- Nicolas to review the current text second (check some GP statements).
- Nicolas to review BioVel and check Galaxy. Other citations to do: TaxaMatch, PESI, WoRMS.
- Complete the references.
Next meeting
Thursday 27 February 2014, 11:00 am for the group.