27.02.2014 BiolDiv
Meeting 27 February 2014, 11:00 am
Google Hangout
Present: Anton, Nicolas, Casey, Edward
Notes
Meeting in Rome
It seems that there will be support for Edward to attend the iMarine meeting in Rome after all. Edward is waiting for confirmation of this news and the precise agenda (is being drafted by Marc Taconet), but will already look into flights and accommodation.
There will probably only be 20 minutes in the programme allocated to BiOnym: 10 minutes presentation, 10 minutes discussion. Anton proposes the following outline for the presentation:
- First half: Fabio on general matching, including overview of potential applications, and developments in FAO
- Second half: Edward on specifics for BiOnym
Fabio and Edward to contact each other ASAP. GP is available, also through skype, to help with screenshots or other information.
User Interface
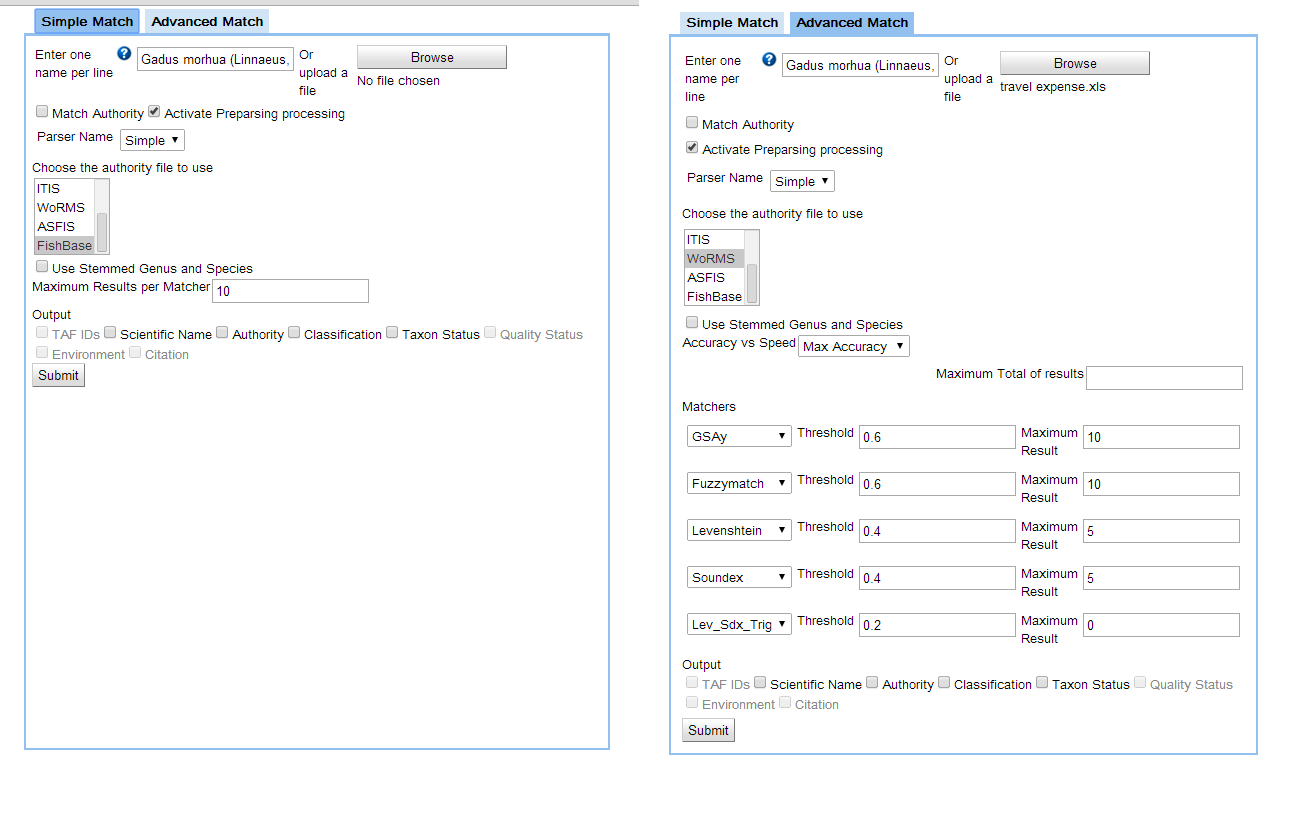
Casey shared the latest version of the BiOnym user interface, on https://www.dropbox.com/s/lx8fwre6dq0zgxd/bionym_interface.png.
Screenshot of the draft BiOnym user interface
There is now a section dealing with output, but this should be expanded. We could consider creating a third tab, next to 'advanced' and 'simple match', to give us a bit more space to play around. There should be several ways for the user to access the results:
- directly, displayed in the user interface (in a separate window?) for small data sets that can be calculated in a time acceptible for a waiting time for a browser
- delayed for larger data sets/more complicated calculations
- either by displaying a link to an ftp file as soon as the output as generated (and a notification email with the same link); this is the method used by NODC in Silver Spring, in WODSelect (where queries also routinely are taking too long for the user to be expected to keep his browser open and wait for the results to be returned)
- or choosing a location in the statistical manager (again with an email notification). From the Data Space, the file can then be transferred to the iMarine Workspace, and used as input for other processes, and shared with other users and/or groups. More links with Data Manager to be investigated.
- LINO COMMENT: Why you are not thinking to exploit the Workspace? The Workspace offers a very simply-to-use Java interface through which it is possible to store a file and get a URL to access it. The output could be saved in the workspace by the Statistical Manager. The exploitation of an FTP server seems to me out of scope in this case for a number of good reasons that I can easily explain if needed.
The uploaded file is transformed into a table. It is possible to avoid the current full upload process by making it transparent to the user, unless he clicks on a tick box aside the browse box. In a way it is to make transparent to the common user the file handling in the infrastructure. Maybe check if we can handle the table through the Data Manager in a later stage.
There was some discussion as to whether ASFIS should be treated the same way as the other taxonomic authority files, as ASFIS is a bit different, and much narrower in its scope. In view of the special role within FAO, it is important to leave ASFIS as one of the authority files to choose from, but we should probably rename 'authority file' to 'reference file'.
Article
Some action points:
- Expand the intro with real examples to make it more concrete. Examples: misspellings from OBIS; different interpretation of vernacular names in different regions, even for the same language (e.g. UK English vs US English: inversion of interpretation of 'Shrimp' and 'Prawn')
- Complete Section 2 (Nicolas) on Taxamatch and Biovel.
- All, check section 3.1 that should be the outline of the following sections.
For the format: a possibility is to write a comprehensive document now, and to publish it as a CNR Publication through the PUMA system: there the report would be available and downloadable, also from outside CNR. From there, we can summarise the information in different ways, for different audiences. This way we should create at least two articles, one for informatics, one in biodiversity, referring to each other and to the complete CNR report.
- LINO COMMENT: I endorse this proposal.
Background for the users' guide can also be included in the CNR Report. Details on the operation of the user interface are better on a wiki.
For publication of scientific articles, following were mentioned (but not really discussed in epth):
- PLoS One is seen as too expensive
- for biodiversity journals
- J. Linn. Soc. (but not very likely)
- Biodiversity Informatics (should be within scope; must check citation index)
- Taxon
Validation
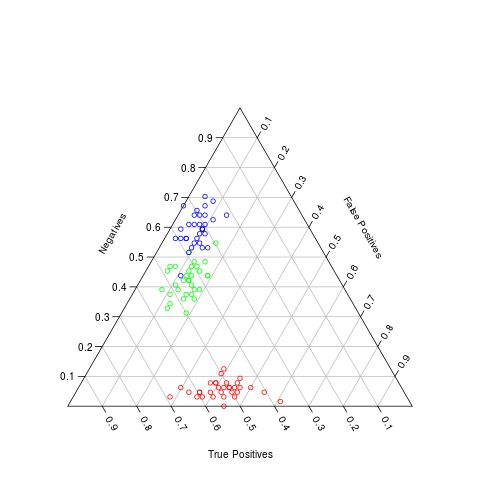
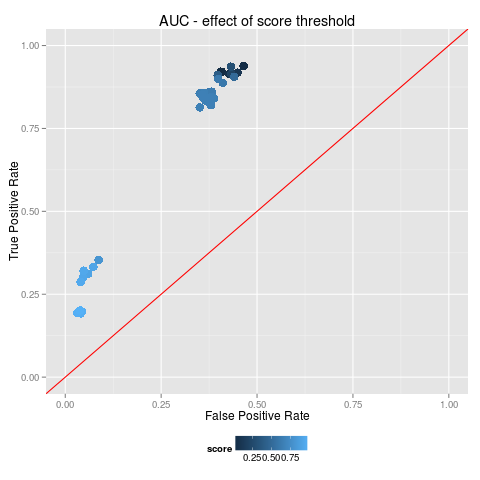
GP and Edward have been working on the technical validation and quality assurance of the output of BiOnym. New plots are available: Triax diagrams and AUC/ROC curves. Both of these are now used to illustrate the performance of the full workflow; now they have been use to investigate the performance as a a function of the threshold for the 'score' of a match. GP will run some additional experiments and Edward will use the results to compare different workflows (e.g. our full BiOnym vs only Levenshtein or Trigram; BiOnym vs TaxaMatch or others).
Triax diagram; example plot relevant to technical validation/Quality assessment of BiOnym output
AUC/ROC curve; example plot relevant to technical validation/Quality assessment of BiOnym output
Apart from the technical validation, we also need validation by the community. GP reported that he got very good and enthusiastic feedback from Philippe Couby on his presentation there of the iMarine infrastructure, including BiOnym.
Next meetings
Thursday 27 February, 11:00 am for the group - conditional on availability of enough participants.