Difference between revisions of "19.09.2013 BiolDiv"
m |
m |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 5: | Line 5: | ||
'''Notes''' | '''Notes''' | ||
| + | ===Noon=== | ||
| + | The main topic of the meeting was BiOnym, and what should be done at short notice to have a working demonstration version of the system, which is still in concept at this point. | ||
| + | In BiOnym as it was envisaged originally, the user can choose between different reference list to try and resolve his/her names. Some of these lists are the reference lists that were uploaded in the iMarine Infrastructure already (CoL, WoRMS, ITIS, FishBase…); in the original concept the user would also have the opportunity to upload his own reference list. It was decided that uploading user-defined reference lists should be postponed. | ||
| + | |||
| + | Reference lists can be accessed either through web services on the fly, or the iMarine infrastructure could store a cache. Using a reference list in BiOnym would probably involve some preparation of the reference list – such as making sure the format of the reference list is understood by our matching process, and calculating some extra fields such as soundex. For technical reasons, caching and creating these extra fields in a ‘pre-processing’ of the reference lists might be the best solution. This pre-processing could be defined as a separate activity (service? tool?) in the infrastructure. | ||
| + | |||
| + | Darwin Core Archive was discussed as a possible format for the pre-processed reference lists. The core of DwCA is a CSV text file with the data; other files are documenting the exact nature of the columns in the data file. Fabio’s SpeciMEn.jar works with reference lists that are incorporated in the .jar file, using a CSV format to store the data and the extra fields. Lino made exports from WoRMS available as an example. | ||
| + | |||
| + | We need a technical discussion about exactly how to implement Fabio’s matching tool in the iMarine infrastructure. Fabio is open to assist with any solution that is most convenient for the infrastructure. To be continued between FAO and CNR. | ||
| + | |||
| + | The overall structure of the application was discussed. The whole process is dependent on the availability of the reference lists – see above. The process of ‘matching’ would start with the definition of the workflow. In the initialization process, end users define which, and in which order, and with what settings, a number of ‘switches’ (Edward’s term) or ‘matchers’ (Fabio’s term) are applied. He/she chooses a reference list (or, in future, uploads his own reference list and applies the reference list pre-processing on it), and uploads the data he wants to match/test. The test data go to a pre-processing step, consisting of a ‘cleaner’ (stripping extraneous information such as ‘cfr.’, ‘aff.’; harmonizing ‘var’, ‘v.’… to ‘var.’…) and ‘parser’ (atomizing the complete string in its constituents such as genus name, specific epitheton, authority, author year…). From there the data go to the series of switches as defined by the user while initializing the process. In each switch, some names are sent to the post-processing, others are sent on to the next switch. For the post-processing, several options should be defined – such as calculating performance statistics in an experimental run of the matching system; presenting the end user with a list of choices to be made in case a best match could not be made automatically; adding classification, rank and synonymy information to the matched names… | ||
| + | |||
| + | As there still seems to be some confusion over the concept, and as some of the documentation on the iMarine Wiki and Workspace seems to be overlooked, Edward will produce an overview document with links to relevant documentation on the iMarine web sites. [Now available on [[[[http://goo.gl/2ITOdJ]]]] | ||
| + | |||
| + | Integrating the different lines of work within iMarine that touch on BiOnym/name matching is long overdue. We urgently need to resolve this. We should have a discussion involving also Casey and Nicolas as soon as possible. | ||
| + | |||
| + | ===Afternoon=== | ||
We discussed the taxon name matcher BiOnym a bit further, following up on issues that were raised in the noon meeting. We agree that it is important that the different activity streams come together. We don’t know exactly how; Edward’s problem is that he doesn’t know much about Casey’s work so it’s difficult for him to suggest a way forward. GP will pass on some instruction on how to explore what Casey has done on gCube; hopefully this will assist in finding a way forward. We will explore whether Casey’s work can be transformed into a plug-in in Fabio’s .jar, or, alternatively, the output of Casey’s procedure can be merged with Fabio’s. To be continued. | We discussed the taxon name matcher BiOnym a bit further, following up on issues that were raised in the noon meeting. We agree that it is important that the different activity streams come together. We don’t know exactly how; Edward’s problem is that he doesn’t know much about Casey’s work so it’s difficult for him to suggest a way forward. GP will pass on some instruction on how to explore what Casey has done on gCube; hopefully this will assist in finding a way forward. We will explore whether Casey’s work can be transformed into a plug-in in Fabio’s .jar, or, alternatively, the output of Casey’s procedure can be merged with Fabio’s. To be continued. | ||
| Line 14: | Line 31: | ||
Edward will try and create a kind of a ‘prototype’ in R illustrating the whole BiOnym approach; where possible, he’ll put the work on postgres (for many things I have a choice of having things done by R or by PostgreSQL; in fact much of the calculations of the performance statistics are implemented in both, to cross-validate results while developing). The PostgreSQL can be re-used directly by calling the statements from Java. | Edward will try and create a kind of a ‘prototype’ in R illustrating the whole BiOnym approach; where possible, he’ll put the work on postgres (for many things I have a choice of having things done by R or by PostgreSQL; in fact much of the calculations of the performance statistics are implemented in both, to cross-validate results while developing). The PostgreSQL can be re-used directly by calling the statements from Java. | ||
| − | Other topics | + | |
| + | ==Other topics== | ||
Edward extracted inferred absences for cod (Gadus morhua) from the OBIS database; this is done on the basis of datasets which have survey information, looking for a well-defined and limited number of species. Datasets of this type where cod has been recorded clearly have cod on their monitoring list; if there are records in this dataset for other species, but not for cod, this can be inferred to be an absence point for cod. A script selecting what are most probably relevant datasets, and then selecting cod presence and inferred absence points was developed by Edward and shared with GP. Apart from selecting relevant points, the script also plots the data. The R script is now also placed on the iMarine workspace (http://goo.gl/A3UUDX). | Edward extracted inferred absences for cod (Gadus morhua) from the OBIS database; this is done on the basis of datasets which have survey information, looking for a well-defined and limited number of species. Datasets of this type where cod has been recorded clearly have cod on their monitoring list; if there are records in this dataset for other species, but not for cod, this can be inferred to be an absence point for cod. A script selecting what are most probably relevant datasets, and then selecting cod presence and inferred absence points was developed by Edward and shared with GP. Apart from selecting relevant points, the script also plots the data. The R script is now also placed on the iMarine workspace (http://goo.gl/A3UUDX). | ||
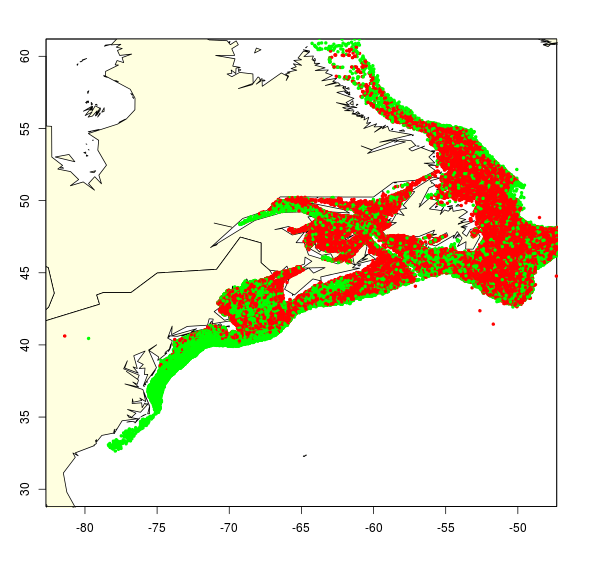
| − | After seeing the dataset on cod presences and calculated/inferred absences, GP is sceptical on whether this will be analysable with Species Distribution Modelling algorithms. There doesn’t seem to be much separating the absence from the presence points (see map; red=presence, green=absence. Many points obscure each other). We will run a number of tests, using bio-oracle combined with ETOPO (or GEBCO) as environmental information. We’ll | + | After seeing the dataset on cod presences and calculated/inferred absences, GP is sceptical on whether this will be analysable with Species Distribution Modelling algorithms. There doesn’t seem to be much separating the absence from the presence points (see map; red=presence, green=absence. Many points obscure each other). We will run a number of tests, using bio-oracle combined with ETOPO (or GEBCO) as environmental information. We’ll compare the predicted range map from presence-only methods (including AquaMaps and Mahalanobis distance) with methods requiring absence data (such as logistic regression, Maxent and random forests). The algorithms are available in the R Package Dismo; we shouldn’t wait till openModeller becomes available in iMarine as this might be too late for us. |
[[File:presence and inferred absence points for cod.png]] | [[File:presence and inferred absence points for cod.png]] | ||
| − | A possible solution to the problem of the ‘closeness’ of presence and inferred absence points is to summarise the data in | + | A possible solution to the problem of the ‘closeness’ of presence and inferred absence points is to summarise the data in squares, with size corresponding to the resolution of Bio-Oracle, and take n(presence)/(n(presence)+n(inferred absence)) as a proxy for abundance (or more formally, for the probability to encounter cod in a sample taken in that square). Then we can use this statistic with SDM methods using abundance. |
The person creating bio-oracle now works at VLIZ. Edward will contact him and ask him whether he can make bio-oracle data available as geo-located layers (e.g. NetCDF-CF GRID), as GP has had some trouble with the ASCII format in which the data is now available. | The person creating bio-oracle now works at VLIZ. Edward will contact him and ask him whether he can make bio-oracle data available as geo-located layers (e.g. NetCDF-CF GRID), as GP has had some trouble with the ASCII format in which the data is now available. | ||
| Line 28: | Line 46: | ||
GP will look into LOF as an alternative to DBScan (as requested by Nicolas in his task list for GP); if it’s implemented in the iMarine infrastructure, it will be via a Java library; GP will send information to Edward on how to call this library from R, as the R implementations of LOF are in rather obscure packages. | GP will look into LOF as an alternative to DBScan (as requested by Nicolas in his task list for GP); if it’s implemented in the iMarine infrastructure, it will be via a Java library; GP will send information to Edward on how to call this library from R, as the R implementations of LOF are in rather obscure packages. | ||
| − | There are still several articles Edward should comment on, including the paper led by Leonardo. For this paper, we will look for another potential journal, as PLoS One doesn’t seem to be best suited. Ecological Informatics (which | + | There are still several articles Edward should comment on, including the paper led by Leonardo. For this paper, we will look for another potential journal, as PLoS One doesn’t seem to be best suited. Ecological Informatics (which Edward doesn’t know) and Ecography (Edward will look for the instructions to authors) are the two we’ll explore now. |
Latest revision as of 10:20, 26 September 2013
Meeting 19 September 2013, 12 noon
Participants: Anton, Lino, GP, Fabio, Edward; continued in the afternoon with only GP and Edward present
Notes
Noon
The main topic of the meeting was BiOnym, and what should be done at short notice to have a working demonstration version of the system, which is still in concept at this point. In BiOnym as it was envisaged originally, the user can choose between different reference list to try and resolve his/her names. Some of these lists are the reference lists that were uploaded in the iMarine Infrastructure already (CoL, WoRMS, ITIS, FishBase…); in the original concept the user would also have the opportunity to upload his own reference list. It was decided that uploading user-defined reference lists should be postponed.
Reference lists can be accessed either through web services on the fly, or the iMarine infrastructure could store a cache. Using a reference list in BiOnym would probably involve some preparation of the reference list – such as making sure the format of the reference list is understood by our matching process, and calculating some extra fields such as soundex. For technical reasons, caching and creating these extra fields in a ‘pre-processing’ of the reference lists might be the best solution. This pre-processing could be defined as a separate activity (service? tool?) in the infrastructure.
Darwin Core Archive was discussed as a possible format for the pre-processed reference lists. The core of DwCA is a CSV text file with the data; other files are documenting the exact nature of the columns in the data file. Fabio’s SpeciMEn.jar works with reference lists that are incorporated in the .jar file, using a CSV format to store the data and the extra fields. Lino made exports from WoRMS available as an example.
We need a technical discussion about exactly how to implement Fabio’s matching tool in the iMarine infrastructure. Fabio is open to assist with any solution that is most convenient for the infrastructure. To be continued between FAO and CNR.
The overall structure of the application was discussed. The whole process is dependent on the availability of the reference lists – see above. The process of ‘matching’ would start with the definition of the workflow. In the initialization process, end users define which, and in which order, and with what settings, a number of ‘switches’ (Edward’s term) or ‘matchers’ (Fabio’s term) are applied. He/she chooses a reference list (or, in future, uploads his own reference list and applies the reference list pre-processing on it), and uploads the data he wants to match/test. The test data go to a pre-processing step, consisting of a ‘cleaner’ (stripping extraneous information such as ‘cfr.’, ‘aff.’; harmonizing ‘var’, ‘v.’… to ‘var.’…) and ‘parser’ (atomizing the complete string in its constituents such as genus name, specific epitheton, authority, author year…). From there the data go to the series of switches as defined by the user while initializing the process. In each switch, some names are sent to the post-processing, others are sent on to the next switch. For the post-processing, several options should be defined – such as calculating performance statistics in an experimental run of the matching system; presenting the end user with a list of choices to be made in case a best match could not be made automatically; adding classification, rank and synonymy information to the matched names…
As there still seems to be some confusion over the concept, and as some of the documentation on the iMarine Wiki and Workspace seems to be overlooked, Edward will produce an overview document with links to relevant documentation on the iMarine web sites. [Now available on [[[[1]]]]
Integrating the different lines of work within iMarine that touch on BiOnym/name matching is long overdue. We urgently need to resolve this. We should have a discussion involving also Casey and Nicolas as soon as possible.
Afternoon
We discussed the taxon name matcher BiOnym a bit further, following up on issues that were raised in the noon meeting. We agree that it is important that the different activity streams come together. We don’t know exactly how; Edward’s problem is that he doesn’t know much about Casey’s work so it’s difficult for him to suggest a way forward. GP will pass on some instruction on how to explore what Casey has done on gCube; hopefully this will assist in finding a way forward. We will explore whether Casey’s work can be transformed into a plug-in in Fabio’s .jar, or, alternatively, the output of Casey’s procedure can be merged with Fabio’s. To be continued.
GP will study Edward’s R script to investigate calling Fabio’s jar from R. The .jar, being java, is OS independent. The jar calls an OS-native helper program (compiled from C) to calculate Levenshtein distance. Depending on the OS on which this will run, we’ll have to install one of the libraries available on http://goo.gl/smYDh0; the native library is not essential, only speeds up calculations.
The R scripts to analyse performance of matching procedures now lives on the iMarine/d4Science SVN (https://svn.d4science.research-infrastructures.eu/gcube/trunk/data-analysis/onym/imarine). The scripts will be duplicated to the workspace.
The abstract for the presentation of the TDWG meeting is considered final. Edward will contact Yde to see what the next steps are – do we need to register; which session will this go in…
Edward will try and create a kind of a ‘prototype’ in R illustrating the whole BiOnym approach; where possible, he’ll put the work on postgres (for many things I have a choice of having things done by R or by PostgreSQL; in fact much of the calculations of the performance statistics are implemented in both, to cross-validate results while developing). The PostgreSQL can be re-used directly by calling the statements from Java.
Other topics
Edward extracted inferred absences for cod (Gadus morhua) from the OBIS database; this is done on the basis of datasets which have survey information, looking for a well-defined and limited number of species. Datasets of this type where cod has been recorded clearly have cod on their monitoring list; if there are records in this dataset for other species, but not for cod, this can be inferred to be an absence point for cod. A script selecting what are most probably relevant datasets, and then selecting cod presence and inferred absence points was developed by Edward and shared with GP. Apart from selecting relevant points, the script also plots the data. The R script is now also placed on the iMarine workspace (http://goo.gl/A3UUDX).
After seeing the dataset on cod presences and calculated/inferred absences, GP is sceptical on whether this will be analysable with Species Distribution Modelling algorithms. There doesn’t seem to be much separating the absence from the presence points (see map; red=presence, green=absence. Many points obscure each other). We will run a number of tests, using bio-oracle combined with ETOPO (or GEBCO) as environmental information. We’ll compare the predicted range map from presence-only methods (including AquaMaps and Mahalanobis distance) with methods requiring absence data (such as logistic regression, Maxent and random forests). The algorithms are available in the R Package Dismo; we shouldn’t wait till openModeller becomes available in iMarine as this might be too late for us.
A possible solution to the problem of the ‘closeness’ of presence and inferred absence points is to summarise the data in squares, with size corresponding to the resolution of Bio-Oracle, and take n(presence)/(n(presence)+n(inferred absence)) as a proxy for abundance (or more formally, for the probability to encounter cod in a sample taken in that square). Then we can use this statistic with SDM methods using abundance.
The person creating bio-oracle now works at VLIZ. Edward will contact him and ask him whether he can make bio-oracle data available as geo-located layers (e.g. NetCDF-CF GRID), as GP has had some trouble with the ASCII format in which the data is now available.
GP will look into LOF as an alternative to DBScan (as requested by Nicolas in his task list for GP); if it’s implemented in the iMarine infrastructure, it will be via a Java library; GP will send information to Edward on how to call this library from R, as the R implementations of LOF are in rather obscure packages.
There are still several articles Edward should comment on, including the paper led by Leonardo. For this paper, we will look for another potential journal, as PLoS One doesn’t seem to be best suited. Ecological Informatics (which Edward doesn’t know) and Ecography (Edward will look for the instructions to authors) are the two we’ll explore now.